Mycoplasma laboratorium
| Mycoplasma laboratorium | |
|---|---|
| Scientific classification | |
| Domain: | Bacteria |
| Phylum: | Mycoplasmatota |
| Class: | Mollicutes |
| Order: | Mycoplasmatales |
| Family: | Mycoplasmataceae |
| Genus: | Mycoplasma |
| Species: | |
| Subspecies: | M. m. JCVI-syn1.0
|
| Trinomial name | |
| Mycoplasma mycoides JCVI-syn1.0 Gibson et al., 2010[a 1]
| |
| Synonyms[a 2] | |
|
Mycoplasma laboratorium Reich, 2000 | |
Mycoplasma laboratorium or Synthia[b 1] refers to a synthetic strain of bacterium. The project to build the new bacterium has evolved since its inception. Initially the goal was to identify a minimal set of genes that are required to sustain life from the genome of Mycoplasma genitalium, and rebuild these genes synthetically to create a "new" organism. Mycoplasma genitalium was originally chosen as the basis for this project because at the time it had the smallest number of genes of all organisms analyzed. Later, the focus switched to Mycoplasma mycoides and took a more trial-and-error approach.[b 2]
To identify the minimal genes required for life, each of the 482 genes of M. genitalium was individually deleted and the viability of the resulting mutants was tested. This resulted in the identification of a minimal set of 382 genes that theoretically should represent a minimal genome.[a 3] In 2008 the full set of M. genitalium genes was constructed in the laboratory with watermarks added to identify the genes as synthetic.[b 3][a 4] However M. genitalium grows extremely slowly and M. mycoides was chosen as the new focus to accelerate experiments aimed at determining the set of genes actually needed for growth.[b 4]
In 2010, the complete genome of M. mycoides was successfully synthesized from a computer record and transplanted into an existing cell of Mycoplasma capricolum that had had its DNA removed.[b 5] It is estimated that the synthetic genome used for this project cost US$40 million and 200 man-years to produce.[b 4] The new bacterium was able to grow and was named JCVI-syn1.0, or Synthia. After additional experimentation to identify a smaller set of genes that could produce a functional organism, JCVI-syn3.0 was produced, containing 473 genes.[b 2] 149 of these genes are of unknown function.[b 2] Since the genome of JCVI-syn3.0 is novel, it is considered the first truly synthetic organism.
Minimal genome project
The production of Synthia is an effort in synthetic biology at the J. Craig Venter Institute by a team of approximately 20 scientists headed by Nobel laureate Hamilton Smith and including DNA researcher Craig Venter and microbiologist Clyde A. Hutchison III. The overall goal is to reduce a living organism to its essentials and thus understand what is required to build a new organism from scratch.[a 3] The initial focus was the bacterium M. genitalium, an obligate intracellular parasite whose genome consists of 482 genes comprising 582,970 base pairs, arranged on one circular chromosome (at the time the project began, this was the smallest genome of any known natural organism that can be grown in free culture). They used transposon mutagenesis to identify genes that were not essential for the growth of the organism, resulting in a minimal set of 382 genes.[a 3] This effort was known as the Minimal Genome Project.[a 5]
Choice of organism
Mycoplasma
Mycoplasma is a genus of bacteria of the class Mollicutes in the division Mycoplasmatota (formerly Tenericutes), characterised by the lack of a cell wall (making it Gram negative) due to its parasitic or commensal lifestyle. In molecular biology, the genus has received much attention, both for being a notoriously difficult-to-eradicate contaminant in mammalian cell cultures (it is immune to beta-lactams and other antibiotics),[a 6] and for its potential uses as a model organism due to its small genome size.[a 7] The choice of genus for the Synthia project dates to 2000, when Karl Reich coined the phrase Mycoplasma laboratorium.[a 2]
Other organisms with small genomes
As of 2005, Pelagibacter ubique (an α-proteobacterium of the order Rickettsiales) has the smallest known genome (1,308,759 base pairs) of any free living organism and is one of the smallest self-replicating cells known. It is possibly the most numerous bacterium in the world (perhaps 1028 individual cells) and, along with other members of the SAR11 clade, are estimated to make up between a quarter and a half of all bacterial or archaeal cells in the ocean.[a 8] It was identified in 2002 by rRNA sequences and was fully sequenced in 2005.[a 9] It is extremely hard to cultivate a species which does not reach a high growth density in lab culture.[a 10][a 11] Several newly discovered species have fewer genes than M. genitalium, but are not free-living: many essential genes that are missing in Hodgkinia cicadicola, Sulcia muelleri, Baumannia cicadellinicola (symbionts of cicadas) and Carsonella ruddi (symbiote of hackberry petiole gall psyllid, Pachypsylla venusta[a 12]) may be encoded in the host nucleus.[a 13] The organism with the smallest known set of genes as of 2013 is Nasuia deltocephalinicola, an obligate symbiont. It has only 137 genes and a genome size of 112 kb.[a 14][b 6]
| species name | number of genes | size (Mbp) |
|---|---|---|
| Candidatus Hodgkinia cicadicola Dsem [1] | 169 | 0.14 |
| Candidatus Carsonella ruddii PV [2] | 182 | 0.16 |
| Candidatus Sulcia muelleri GWSS [3] | 227 | 0.25 |
| Candidatus Sulcia muelleri SMDSEM [4] | 242 | 0.28 |
| Buchnera aphidicola str. Cinara cedri [5] | 357 | 0.4261 |
| Mycoplasma genitalium G37 [6] | 475 | 0.58 |
| Candidatus Phytoplasma mali [7] | 479 | 0.6 |
| Buchnera aphidicola str. Baizongia pistaciae [8] | 504 | 0.6224 |
| Nanoarchaeum equitans Kin4-M [9] | 540 | 0.49 |
Techniques
Several laboratory techniques had to be developed or adapted for the project, since it required synthesis and manipulation of very large pieces of DNA.
Bacterial genome transplantation
In 2007, Venter's team reported that they had managed to transfer the chromosome of the species Mycoplasma mycoides to Mycoplasma capricolum by:
- isolating the genome of M. mycoides: gentle lysis of cells trapped in agar—molten agar mixed with cells and left to form a gel—followed by pulse field gel electrophoresis and the band of the correct size (circular 1.25Mbp) being isolated;
- making the recipient cells of M. capricolum competent: growth in rich media followed starvation in poor media where the nucleotide starvation results in inhibition of DNA replication and change of morphology; and
- polyethylene glycol-mediated transformation of the circular chromosome to the DNA-free cells followed by selection.[a 15]
The term transformation is used to refer to insertion of a vector into a bacterial cell (by electroporation or heatshock). Here, transplantation is used akin to nuclear transplantation.
Bacterial chromosome synthesis
In 2008 Venter's group described the production of a synthetic genome, a copy of M. genitalium G37 sequence L43967, by means of a hierarchical strategy:[a 16]
- Synthesis → 1kbp: The genome sequence was synthesized by Blue Heron in 1,078 1080bp cassettes with 80bp overlap and NotI restriction sites (inefficient but infrequent cutter).
- Ligation → 10kbp: 109 groups of a series of 10 consecutive cassettes were ligated and cloned in E. coli on a plasmid and the correct permutation checked by sequencing.
- Multiplex PCR → 100kbp: 11 Groups of a series of 10 consecutive 10kbp assemblies (grown in yeast) were joined by multiplex PCR, using a primer pair for each 10kbp assembly.
- Isolation and recombination → secondary assemblies were isolated, joined and transformed into yeast spheroplasts without a vector sequence (present in assembly 811-900).
The genome of this 2008 result, M. genitalium JCVI-1.0, is published on GenBank as CP001621.1. It is not to be confused with the later synthetic organisms, labelled JCVI-syn, based on M. mycoides.[a 16]
Synthetic genome
In 2010 Venter and colleagues created Mycoplasma mycoides strain JCVI-syn1.0 with a synthetic genome.[a 1] Initially the synthetic construct did not work, so to pinpoint the error—which caused a delay of 3 months in the whole project[b 4]—a series of semi-synthetic constructs were created. The cause of the failure was a single frameshift mutation in DnaA, a replication initiation factor.[a 1]
The purpose of constructing a cell with a synthetic genome was to test the methodology, as a step to creating modified genomes in the future. Using a natural genome as a template minimized the potential sources of failure. Several differences are present in Mycoplasma mycoides JCVI-syn1.0 relative to the reference genome, notably an E.coli transposon IS1 (an infection from the 10kb stage) and an 85bp duplication, as well as elements required for propagation in yeast and residues from restriction sites.[a 1]
There has been controversy over whether JCVI-syn1.0 is a true synthetic organism. While the genome was synthesized chemically in many pieces, it was constructed to match the parent genome closely and transplanted into the cytoplasm of a natural cell. DNA alone cannot create a viable cell: proteins and RNAs are needed to read the DNA, and lipid membranes are required to compartmentalize the DNA and cytoplasm. In JCVI-syn1.0 the two species used as donor and recipient are of the same genus, reducing potential problems of mismatches between the proteins in the host cytoplasm and the new genome.[a 17] Paul Keim (a molecular geneticist at Northern Arizona University in Flagstaff) noted that "there are great challenges ahead before genetic engineers can mix, match, and fully design an organism's genome from scratch".[b 4]
Watermarks

A much publicized feature of JCVI-syn1.0 is the presence of watermark sequences. The 4 watermarks (shown in Figure S1 in the supplementary material of the paper[a 1]) are coded messages written into the DNA, of length 1246, 1081, 1109 and 1222 base pairs respectively. These messages did not use the standard genetic code, in which sequences of 3 DNA bases encode amino acids, but a new code invented for this purpose, which readers were challenged to solve.[b 7] The content of the watermarks is as follows:
- Watermark 1: an HTML script which reads to a browser as text congratulating the decoder, and instructions on how to email the authors to prove the decoding.
- Watermark 2: a list of authors and a quote from James Joyce: "To live, to err, to fall, to triumph, to recreate life out of life".
- Watermark 3: more authors and a quote from Robert Oppenheimer (uncredited): "See things not as they are, but as they might be".
- Watermark 4: more authors and a quote from Richard Feynman: "What I cannot build, I cannot understand".
JCVI-syn3.0
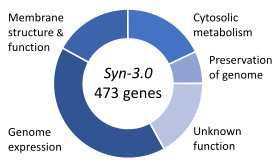
In 2016, the Venter Institute used genes from JCVI-syn1.0 to synthesize a smaller genome they call JCVI-syn3.0, that contains 531,560 base pairs and 473 genes.[b 8] In 1996, after comparing M. genitalium with another small bacterium Haemophilus influenzae, Arcady Mushegian and Eugene Koonin had proposed that there might be a common set of 256 genes which could be a minimal set of genes needed for viability.[b 9][a 19] In this new organism, the number of genes can only be pared down to 473, 149 of which have functions that are completely unknown.[b 9] As of 2022 the unknown set has been narrowed to about 100.[b 10] In 2019 a complete computational model of all pathways in Syn3.0 cell was published, representing the first complete in silico model for a living minimal organism.[a 20]
Concerns and controversy
Reception
On Oct 6, 2007, Craig Venter announced in an interview with UK's The Guardian newspaper that the same team had synthesized a modified version of the single chromosome of Mycoplasma genitalium chemically. The synthesized genome had not yet been transplanted into a working cell. The next day the Canadian bioethics group, ETC Group issued a statement through their representative, Pat Mooney, saying Venter's "creation" was "a chassis on which you could build almost anything. It could be a contribution to humanity such as new drugs or a huge threat to humanity such as bio-weapons". Venter commented "We are dealing in big ideas. We are trying to create a new value system for life. When dealing at this scale, you can't expect everybody to be happy."[b 11]
On May 21, 2010, Science reported that the Venter group had successfully synthesized the genome of the bacterium Mycoplasma mycoides from a computer record and transplanted the synthesized genome into the existing cell of a Mycoplasma capricolum bacterium that had had its DNA removed. The "synthetic" bacterium was viable, i.e. capable of replicating.[b 1] Venter described it as "the first species.... to have its parents be a computer".[b 12]
The creation of a new synthetic bacterium, JCVI-3.0 was announced in Science on March 25, 2016. It has only 473 genes. Venter called it “the first designer organism in history” and argued that the fact that 149 of the genes required have unknown functions means that "the entire field of biology has been missing a third of what is essential to life".[a 21]
Press coverage
The project received a large amount of coverage from the press due to Venter's showmanship, to the degree that Jay Keasling, a pioneering synthetic biologist and founder of Amyris commented that "The only regulation we need is of my colleague's mouth".[b 13]
Utility
Venter has argued that synthetic bacteria are a step towards creating organisms to manufacture hydrogen and biofuels, and also to absorb carbon dioxide and other greenhouse gases. George M. Church, another pioneer in synthetic biology, has expressed the contrasting view that creating a fully synthetic genome is not necessary since E. coli grows more efficiently than M. genitalium even with all its extra DNA; he commented that synthetic genes have been incorporated into E.coli to perform some of the above tasks.[b 14]
Intellectual property
The J. Craig Venter Institute filed patents for the Mycoplasma laboratorium genome (the "minimal bacterial genome") in the U.S. and internationally in 2006.[b 15][b 16][a 22] The ETC group, a Canadian bioethics group, protested on the grounds that the patent was too broad in scope.[b 17]
Similar projects
From 2002 to 2010, a team at the Hungarian Academy of Science created a strain of Escherichia coli called MDS42, which is now sold by Scarab Genomics of Madison, WI under the name of "Clean Genome. E.coli",[b 18] where 15% of the genome of the parental strain (E. coli K-12 MG1655) were removed to aid in molecular biology efficiency, removing IS elements, pseudogenes and phages, resulting in better maintenance of plasmid-encoded toxic genes, which are often inactivated by transposons.[a 23][a 24][a 25] Biochemistry and replication machinery were not altered.
References
Primary sources
- ^ a b c d e Gibson, D. G.; Glass, J. I.; Lartigue, C.; Noskov, V. N.; Chuang, R.-Y.; Algire, M. A.; Benders, G. A.; Montague, M. G.; Ma, L.; Moodie, M. M.; Merryman, C.; Vashee, S.; Krishnakumar, R.; Assad-Garcia, N.; Andrews-Pfannkoch, C.; Denisova, E. A.; Young, L.; Qi, Z.-Q.; Segall-Shapiro, T. H.; Calvey, C. H.; Parmar, P. P.; Hutchison, C. A.; Smith, H. O.; Venter, J. C. (20 May 2010). "Creation of a Bacterial Cell Controlled by a Chemically Synthesized Genome". Science. 329 (5987): 52–56. Bibcode:2010Sci...329...52G. doi:10.1126/science.1190719. PMID 20488990.
- ^ a b Reich, KA (June 2000). "The search for essential genes". Research in Microbiology. 151 (5): 319–24. doi:10.1016/S0923-2508(00)00153-4. PMID 10919511.
In addition, the difficult genetics in these organisms makes subsequent verification of essentiality by directed knockouts problematic and virtually precludes the possibility of performing a de novo synthesis of 'M. laboratorium', the origin of the attention in the popular press.
- ^ a b c Glass, John I.; Nacyra Assad-Garcia; Nina Alperovich; Shibu Yooseph; Matthew R. Lewis; Mahir Maruf; Clyde A. Hutchison; Hamilton O. Smith; J. Craig Venter (2006-01-10). "Essential genes of a minimal bacterium". Proceedings of the National Academy of Sciences. 103 (2): 425–430. Bibcode:2006PNAS..103..425G. doi:10.1073/pnas.0510013103. PMC 1324956. PMID 16407165.
- ^ Gibson, D. G.; Benders, G. A.; Andrews-Pfannkoch, C.; Denisova, E. A.; Baden-Tillson, H.; Zaveri, J.; Stockwell, T. B.; Brownley, A.; Thomas, D. W. (2008-02-29). "Complete Chemical Synthesis, Assembly, and Cloning of a Mycoplasma genitalium Genome". Science. 319 (5867): 1215–1220. Bibcode:2008Sci...319.1215G. doi:10.1126/science.1151721. ISSN 0036-8075. PMID 18218864. S2CID 8190996.
- ^ Hutchison CA, Montague MG (2002). "Mycoplasmas and the minimal genome concept". Molecular Biology and Pathogenicity of Mycoplasmas (Razin S, Herrmann R, eds.). New York: Kluwer Academic/Plenum. pp. 221–54. ISBN 978-0-306-47287-9.
- ^ Young L, Sung J, Stacey G, Masters JR. "Detection of Mycoplasma in cell cultures". Nat Protoc. 2010 5(5): 929–34. Epub 2010 Apr 22.
- ^ Fraser CM, Gocayne JD, White O, et al. (October 1995). "The minimal gene complement of Mycoplasma genitalium". Science. 270 (5235): 397–403. Bibcode:1995Sci...270..397F. doi:10.1126/science.270.5235.397. PMID 7569993. S2CID 29825758.
- ^ Morris RM, et al. (2002). "SAR11 clade dominates ocean surface bacterioplankton communities". Nature. 420 (6917): 806–10. Bibcode:2002Natur.420..806M. doi:10.1038/nature01240. PMID 12490947. S2CID 4360530.
- ^ Stephen J. Giovannoni, H. James Tripp, et al. (2005). "Genome Streamlining in a Cosmopolitan Oceanic Bacterium". Science. 309 (5738): 1242–1245. Bibcode:2005Sci...309.1242G. doi:10.1126/science.1114057. PMID 16109880. S2CID 16221415.
- ^ Rappé MS, Connon SA, Vergin KL, Giovannoni SL (2002). "Cultivation of the ubiquitous SAR11 marine bacterioplankton clade". Nature. 418 (6898): 630–33. Bibcode:2002Natur.418..630R. doi:10.1038/nature00917. PMID 12167859. S2CID 4352877.
- ^ Tripp HJ, Kitner JB, Schwalbach MS, Dacey JW, Wilhelm LJ, Giovannoni SJ (Apr 10, 2008). "SAR11 marine bacteria require exogenous reduced sulphur for growth". Nature. 452 (7188): 741–4. Bibcode:2008Natur.452..741T. doi:10.1038/nature06776. PMID 18337719. S2CID 205212536.
- ^ Nakabachi, A.; Yamashita, A.; Toh, H.; Ishikawa, H.; Dunbar, H. E.; Moran, N. A.; Hattori, M. (2006). "The 160-Kilobase Genome of the Bacterial Endosymbiont Carsonella". Science. 314 (5797): 267. doi:10.1126/science.1134196. PMID 17038615. S2CID 44570539.
- ^ McCutcheon, J. P.; McDonald, B. R.; Moran, N. A. (2009). "Convergent evolution of metabolic roles in bacterial co-symbionts of insects". Proceedings of the National Academy of Sciences. 106 (36): 15394–15399. Bibcode:2009PNAS..10615394M. doi:10.1073/pnas.0906424106. PMC 2741262. PMID 19706397.
- ^ Nancy A. Moran; Gordon M. Bennett (2014). "The Tiniest Tiny Genomes". Annual Review of Microbiology. 68: 195–215. doi:10.1146/annurev-micro-091213-112901. PMID 24995872.
- ^ Lartigue C, Glass JI, Alperovich N, Pieper R, Parmar PP, Hutchison CA 3rd, Smith HO, Venter JC (Aug 3, 2007). "Genome transplantation in bacteria: changing one species to another". Science. 317 (5838): 632–8. Bibcode:2007Sci...317..632L. CiteSeerX 10.1.1.395.4374. doi:10.1126/science.1144622. PMID 17600181. S2CID 83956478.
{cite journal}: CS1 maint: multiple names: authors list (link) CS1 maint: numeric names: authors list (link) - ^ a b Gibson, B; Clyde A. Hutchison; Cynthia Pfannkoch; J. Craig Venter; et al. (2008-01-24). "Complete Chemical Synthesis, Assembly, and Cloning of a Mycoplasma genitalium Genome". Science. 319 (5867): 1215–20. Bibcode:2008Sci...319.1215G. doi:10.1126/science.1151721. PMID 18218864. S2CID 8190996.
- ^ Povolotskaya, IS; Kondrashov, FA (Jun 2010). "Sequence space and the ongoing expansion of the protein universe". Nature. 465 (7300): 922–6. Bibcode:2010Natur.465..922P. doi:10.1038/nature09105. PMID 20485343. S2CID 4431215.
- ^ Hutchison, Clyde A.; Chuang, Ray-Yuan; Noskov, Vladimir N.; Assad-Garcia, Nacyra; Deerinck, Thomas J.; Ellisman, Mark H.; Gill, John; Kannan, Krishna; Karas, Bogumil J. (2016-03-25). "Design and synthesis of a minimal bacterial genome". Science. 351 (6280): aad6253. Bibcode:2016Sci...351.....H. doi:10.1126/science.aad6253. ISSN 0036-8075. PMID 27013737.
- ^ Arcady R. Mushegian; Eugene V. Koonin (September 1996). "A minimal gene set for cellular life derived by comparison of complete bacterial genomes". Proc. Natl. Acad. Sci. USA. 93 (19): 10268–10273. Bibcode:1996PNAS...9310268M. doi:10.1073/pnas.93.19.10268. PMC 38373. PMID 8816789.
- ^ Breuer, Marian; Earnest, Tyler M.; Merryman, Chuck; Wise, Kim S.; Sun, Lijie; Lynott, Michaela R.; Hutchison, Clyde A.; Smith, Hamilton O.; Lapek, John D.; Gonzalez, David J.; De Crécy-Lagard, Valérie; Haas, Drago; Hanson, Andrew D.; Labhsetwar, Piyush; Glass, John I.; Luthey-Schulten, Zaida (2019). "Essential metabolism for a minimal cell". eLife. 8. doi:10.7554/eLife.36842. PMC 6609329. PMID 30657448.
- ^ Herper, Matthew. "After 20 Year Quest, Biologists Create Synthetic Bacteria With No Extra Genes". Forbes. Retrieved 2019-07-02.
- ^ US Patent Application: 20070122826
- ^ Umenhoffer K, Fehér T, Balikó G, Ayaydin F, Pósfai J, Blattner FR, Pósfai G (2010). "Reduced evolvability of Escherichia coli MDS42, an IS-less cellular chassis for molecular and synthetic biology applications". Microbial Cell Factories. 9: 38. doi:10.1186/1475-2859-9-38. PMC 2891674. PMID 20492662.
- ^ Pósfai G, Plunkett G 3rd, Fehér T, Frisch D, Keil GM, Umenhoffer K, Kolisnychenko V, Stahl B, Sharma SS, de Arruda M, Burland V, Harcum SW, Blattner FR (2006). "Emergent properties of reduced-genome Escherichia coli". Science. 312 (5776): 1044–6. Bibcode:2006Sci...312.1044P. doi:10.1126/science.1126439. PMID 16645050. S2CID 43287314.
{cite journal}: CS1 maint: multiple names: authors list (link) CS1 maint: numeric names: authors list (link) - ^ Kolisnychenko V, Plunkett G 3rd, Herring CD, Fehér T, Pósfai J, Blattner FR, Pósfai G (April 2002). "Engineering a reduced Escherichia coli genome". Genome Res. 12 (4): 640–7. doi:10.1101/gr.217202. PMC 187512. PMID 11932248.
{cite journal}: CS1 maint: multiple names: authors list (link) CS1 maint: numeric names: authors list (link)
Popular press
- ^ a b Roberta Kwok (2010). "Genomics: DNA's master craftsmen". Nature. 468 (7320): 22–5. Bibcode:2010Natur.468...22K. doi:10.1038/468022a. PMID 21048740.
- ^ a b c Callaway, Ewen (2016). "'Minimal' cell raises stakes in race to harness synthetic life". Nature. 531 (7596): 557–558. Bibcode:2016Natur.531..557C. doi:10.1038/531557a. ISSN 0028-0836. PMID 27029256.
- ^ Ball, Philip (2008-01-24). "Genome stitched together by hand". Nature. doi:10.1038/news.2008.522. ISSN 0028-0836.
- ^ a b c d Pennisi E (May 2010). "Genomics. Synthetic genome brings new life to bacterium" (PDF). Science. 328 (5981): 958–9. doi:10.1126/science.328.5981.958. PMID 20488994.
- ^ Katsnelson, Alla (2010-05-20). "Researchers start up cell with synthetic genome". Nature. doi:10.1038/news.2010.253. ISSN 0028-0836.
- ^ Zimmer, Carl (2013-08-23). "And the Genomes Keep Shrinking..." National Geographic. Archived from the original on August 23, 2013.
- ^ Ken Shirriff (2010-06-10). "Using Arc to decode Venter's secret DNA watermark". Ken Shirriff's blog. Retrieved 2010-10-29.
- ^ First Minimal Synthetic Bacterial Cell. Astrobiology Web. March 24, 2016.
- ^ a b Yong, Ed (March 24, 2016). "The Mysterious Thing About a Marvelous New Synthetic Cell".
- ^ Somers, James (7 March 2022). "A Journey to the Center of Our Cells". The New Yorker. New York: Condé Nast.
- ^ Pilkington, Ed (6 Oct 2009). "I am creating artificial life, declares US gene pioneer". The Guardian. London. Retrieved 23 Nov 2012.
- ^ "How scientists made 'artificial life'". BBC News. 2010-05-20. Retrieved 2010-05-21.
- ^ Pollack, Andrew (September 4, 2010). "His Corporate Strategy: The Scientific Method". The New York Times.
- ^ Longest Piece of Synthetic DNA Yet, Scientific American News, 24 January 2008
- ^ "Artificial life: Patent pending", The Economist, June 14, 2007. Retrieved October 7, 2007.
- ^ Roger Highfield, "Man-made microbe 'to create endless biofuel'", Telegraph, June 8, 2007. Retrieved October 7, 2007.
- ^ Ianculovici, Elena (2008-01-15). "The Emerging Field of Synthetic Biology: "Syn" or Salvation?". Science in the News. Retrieved 2019-07-03.
- ^ "Scarab Genomics LLC. Company web site".